About the SDK
Overview
The Seven Bridges Platform allows you to bring your own tools and execute them on the Platform. This is done through our Software Development Kit (SDK) and the process consists of the following steps:
- Create a Docker image containing the tool and its dependencies. Push the image to the Seven Bridges Image Registry.
- Use the tool editor on the Platform to create a description of the tool's functionalities. The description is automatically transcribed into the Common Workflow Language (CWL). This process is also known as wrapping.
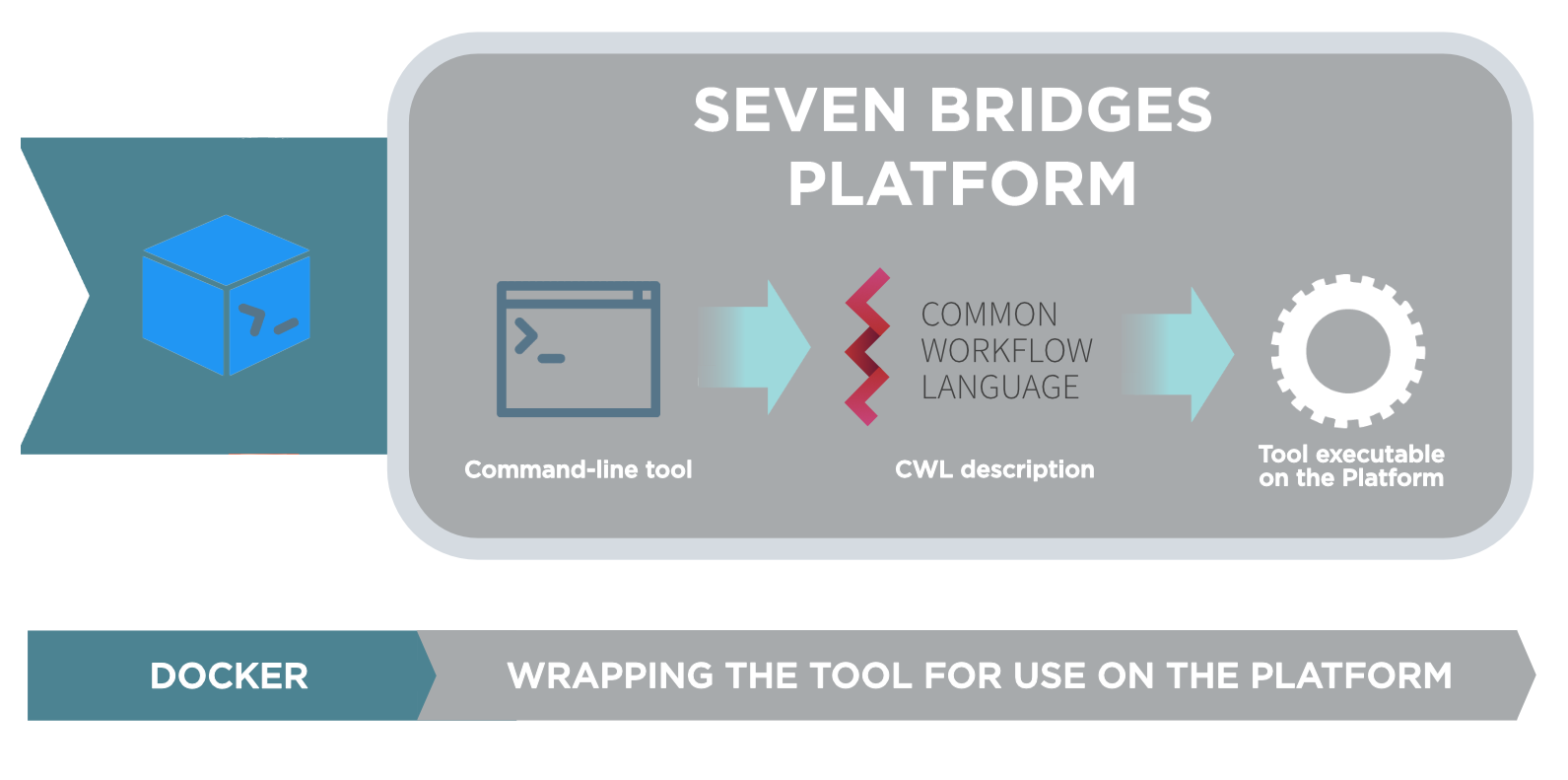
This means that there is no need to reconfigure your existing command line tools to meet any proprietary format. Additionally, the tools remain runnable across a diverse range of infrastructures should you want to use them on different platforms.
A list of common terms and concepts related to bringing your own tools to the Platform is provided in the following sections.
DOCKER
You can use Docker to build and run Docker containers containing your tools, along with their dependencies. Then, you can push snapshots of these containers, called images, to the Seven Bridges Platform Image Registry, which is housed on our computational platform, or to Docker hub – Docker's own image registry. The tools you have installed will be run inside the containers on the Platform.
THE TOOL EDITOR
Having uploaded a Docker image containing your tool to the image registry, you can specify its behavior, including its inputs and outputs, runtime requirements, and execution semantics. The specification is entered using the Tool Editor. It allows the tool to be used on the Seven Bridges Platform to interface with other arbitrary tools.
THE COMMON WORKFLOW LANGUAGE
The specification of your tool that you enter using the Tool Editor will be automatically transcribed into the Common Workflow Language (CWL). This is a community developed, open specification for bioinformatics workflows.
Workflows constructed on the Seven Bridges Platform can also be described using the CWL. This supports reproducibility of workflows in two ways:
1. CWL specifications are exhaustive:
The CWL specifies all configurable details of a workflow execution, right down to each tool's parameterization and the configuration of the environment in which the tools are executed.
This information is provided and stored for every workflow you run on the Seven Bridges Platform. It allows you to easily reference any results obtained, and to provide colleagues with all the information they need to run identical executions.
2. CWL specifications are platform-agnostic:
Since the CWL is an open specification, workflows described using it can be executed on any platform that supports the specification.
To start installing and running your own bioinformatics tools on the Seven Bridges Platform, follow this ten-minute tutorial.
An alternative to the Tool Editor:If you are familiar with the Common Workflow Language you are free to upload your own Common Workflow Language description of the tool's behavior.
Updated 4 months ago
