About datasets
Overview
As a researcher or a clinician you may be interested in accessing publicly available datasets to complement your research and compare findings. As an authorized user you can access these data through the Platform.
These datasets are available on the Seven Bridges sister platform, the Cancer Genomics Cloud, powered by Seven Bridges (CGC), through several data repositories:
- Genomic Data Commons - GDC
- Proteomic Data Commons - PDC
- Cancer Data Commons - CDS
- Integrated Canine Data Commons - ICDC
- The Cancer Imaging Archive - TCIA
Most of these datasets include both Open and Controlled Data. While all data in TCGA and the other datasets is stripped of direct identifiers, DNA information is inherently unique to an individual.
Two types of data access ‘tiers’ have been put in place to balance the desire to make the data as widely available as possible while ensuring that the rights of study participants are well protected. These two access tiers are described below:
Open Data includes information which is not unique to an individual. This includes information such as:
- De-identified clinical and demographic data
- Gene expression data
- Copy number alterations in regions of the genome
- Epigenetic data
- Summaries of data across individuals
Controlled Data includes information which is unique to an individual. This includes most raw data files and some processed data such as:
- Primary sequencing data (BAM and FASTQ files) from DNA, RNA, miRNA or bisulfite sequencing studies
- Raw and processed SNP6 array data
- Raw and processed Exon array data
- Somatic and germ-line mutation calls for an individual (VCF and MAF files)
Once the dbGAP credentials are approved, you will be able to use the data in an analysis, both on CGC and on the Platform. Please note that The Cancer Imaging Archive (TCIA) is public and does not require dbGAP authorization.
User Responsibilities
Seven Bridges is an NIH Trusted Partner, and we've made data security a priority. In addition, users are required to abide by their dbGaP data access requests and the NIH Genomic Data User Code of Conduct, the elements of which are reproduced below:
- Investigator(s) will use requested datasets solely in connection with the research project described in the approved Data Access Request for each dataset;
- Investigator(s) will make no attempt to identify or contact individual participants from whom these data were collected without appropriate approvals from the relevant IRBs;
- Investigator(s) will not distribute these data to any entity or individual beyond those specified in the approved Data Access Request;
- Investigator(s) will adhere to computer security practices that ensure that only authorized individuals can gain access to data files;
- Investigator(s) will not submit for publication or any other form of public dissemination analyses or other reports on work using or referencing NIH datasets prior to the embargo release date listed for the dataset (or dataset version) on dbGaP;
- Investigator(s) acknowledge the Intellectual Property Policies as specified in the Data Use Certification; and,
- Investigator(s) will report any inadvertent data release in accordance with the terms in the Data Use Certification, breach of data security, or other data management incidents contrary to the terms of data access.
Learn more about updating your Data Access Request to list Seven Bridges as the Platform as a Service (PaaS) and include cloud use.
Authenticate and access data
The data above is available through an integration with the Cancer Genomics Cloud (CGC), powered by Seven Bridges.
CGC is a source that allows you to authenticate with dbGaP and CGC and gain access to datasets such as TCGA, CPTAC, and TARGET.
For more information about the CGC, please visit www.cancergenomicscloud.org.
To be able to authenticate and access data, you will first need to create an account on CGC. After registering for a CGC account, you can connect your CGC account to your Seven Bridges Platform account and associate your CGC credentials.
Register for an account on CGC
You can sign up for CGC using your eRA Commons or NIH cit credentials or your email address.
To access Controlled Data on CGC, you need to register with eRA Commons credentials, which have the appropriate data access permissions through dbGaP.
If you use your email address to register, you will only be able to access Open Data.
We encourage you to read the CGC Terms of Use and TCGA Data Use policy to learn more about CGC.
Register using eRA Commons credentials
To register for an account using eRA Commons credentials, follow these steps:
- Access the CGC.
- Click Create an account.
- Click Continue with eRA Commons.
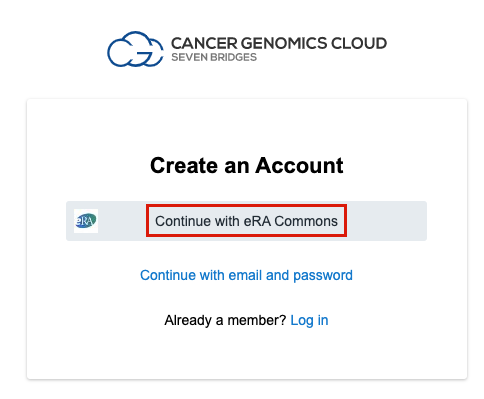
- You are redirected to the iTrust login page, where you should enter your eRA Commons or NIH CIT credentials and click Log in.
- Click Yes, I authorize. You are redirected back to CGC registration form.
- Fill out the registration form.
- Click Proceed to the CGC.
Register using your email address
To register with your email address:
- Access CGC.
- Click Create an account.
- Click Continue with email and password.
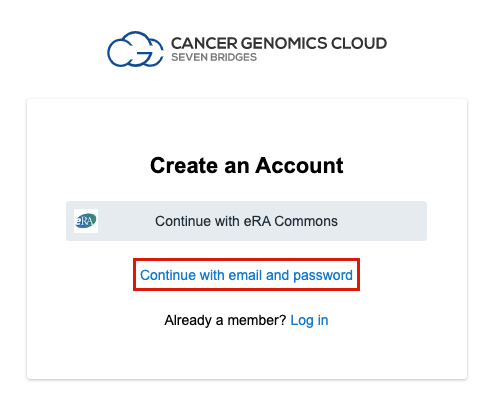
- Fill out the registration form.
- Click Register. CGC will send you an email containing a verification link.
- Open the email from CGC and click Confirm your email.
Connect your CGC account TO your Seven Bridges account
Once you have created a CGC account, you can connect it to your Seven Bridges account. Your CGC credentials will be associated with your Seven Bridges account, and you will be able to access publicly available data right away.
Prerequisites
- Multi-factor authentication has to be enabled on both Seven Bridges Platform and CGC.
- You have to connect your Seven Bridges Platform account to the DRS server (CGC). More on this below.
Connect to the DRS server (CGC)
- Click your username in the upper right corner and then click Account Settings.
- Click the Dataset Access tab.
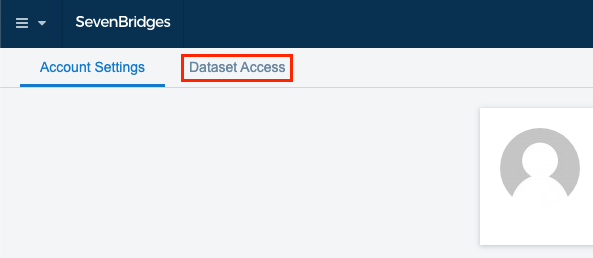
- Locate "Cancer Genomics Cloud Powered by Seven Bridges" section and click Connect.
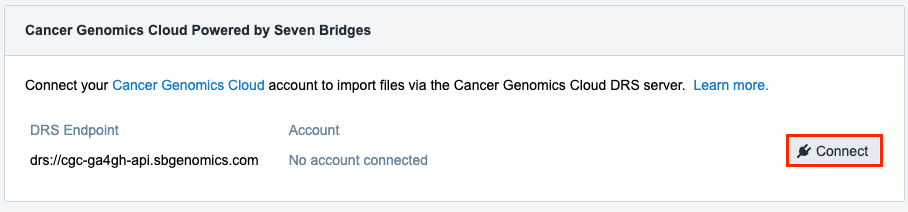
- The following pop-up is displayed.
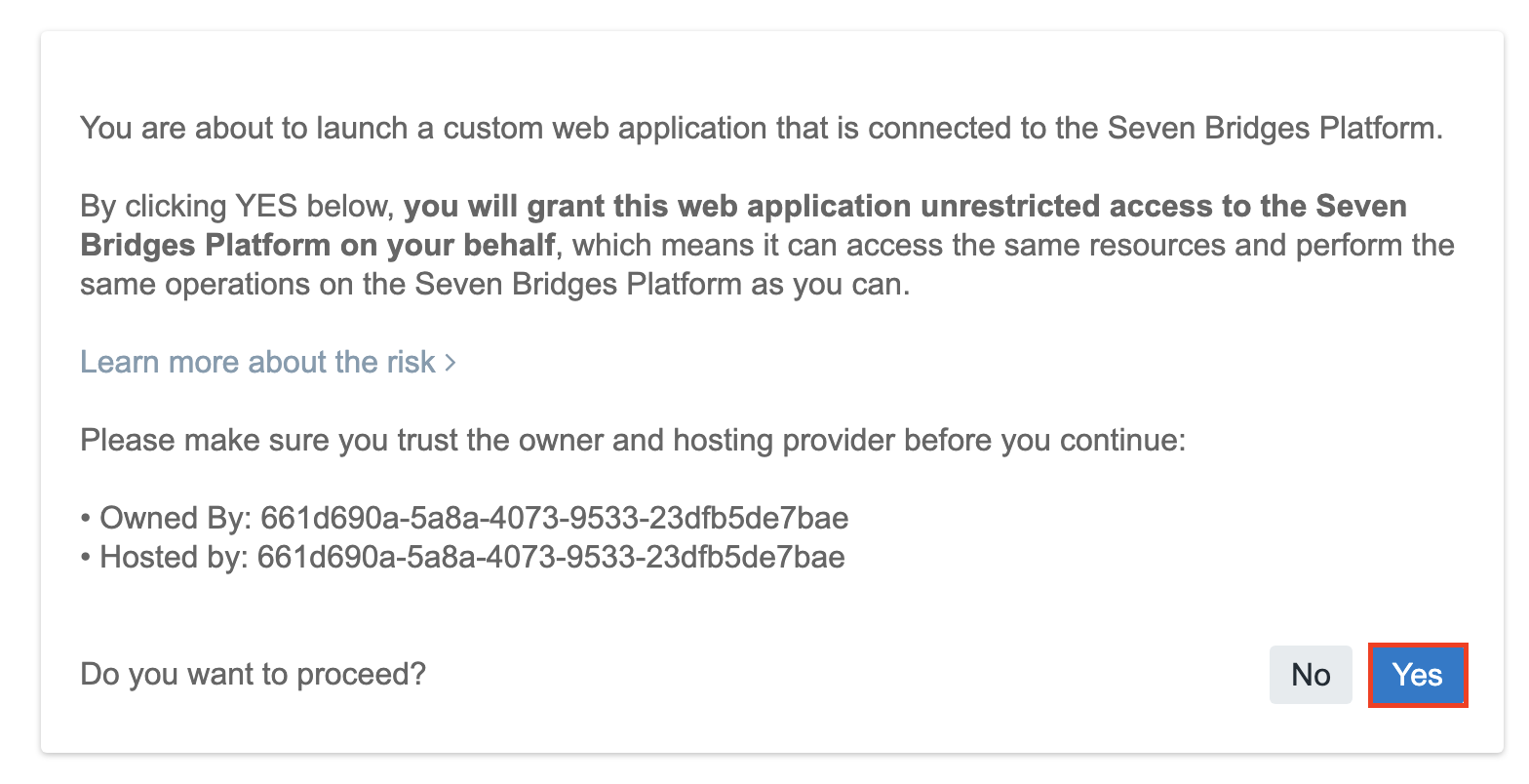
- Click Yes to complete the procedure.
Once completed, your Seven Bridges Platform account is connected to the CGC and you can start importing files using the import via DRS feature.
NoteThis connection will automatically expire in 30 days. You can disconnect your accounts at any time.
Disconnect your account from CGC
Follow the procedure below to disconnect your account from the CGC. Please keep in mind that you will no longer be able to import files. In addition, the files that have been previously imported will no longer be available for use.
- Click your username in the upper right corner and choose option Account Settings.
- Click the Dataset Access tab.
- Find the section for the environment you want to disconnect.
- Click the ellipsis menu and select Deactivate.
Your account is now disconnected from the CGC.
Bring CGC data to Seven Bridges Platform
You can bring your CGC files with associated metadata to the Seven Bridges Platform in just a few steps. Please note that the file content is only referenced, and not actually transferred to Seven Bridges Platform.
Steps on the CGC
- Use available CGC features to explore available datasets such as Data Browser (GDC, TCIA), Cancer Data Service Explorer (CDS), etc.
- OR use external data portals to browse datasets and bring them to the CGC, such as PDC Portal, ICDC Portal
- Import/copy data to the CGC project
- Generate DRS Manifest
- Download DRS Manifest to your local machine
Steps on the Seven Bridges Platform
Follow instructions for importing files from a DRS server.
What type of data can you access?
Once you register for a CGC account, you will have access to various data based on your approved data access.
Open Data access
All Platform users can access Open Data as soon as they create and connect their CGC account and agree to the use certifications as mentioned above.
Controlled Data access
Researchers requiring access to Controlled Data for their studies are required to obtain an approved Data Access Request through dbGaP and to agree to the respective data agreements and publication guidelines.
If you are either a PI or a downloader in an approved dbGaP application, be sure to list Seven Bridges as the Platform as a Service (PaaS) in your dbGaP application.
Updated 4 months ago
