Overview
BETAThis page documents the BETA version of the Variant Browser app with features that are limited in scope.
Please test it using the provided demo VCF files and send your feedback and comments to [email protected]..
The Variant Browser app allows you to analyze and interpret annotated VCF files. Variants are annotated with external databases such as Uniprot, Ensembl, PubMed, Cosmic, dbSNP, Reactome and Swissprot. Each variant links back to the original source where you can get more information. Currently, the app works with WES and gene panels, supporting either single VCF files or trio VCFs.
Variant Browser contains many filters which will allow you to filter by Phred-scaled quality score, variant allele fraction, population allele frequencies (from GnomAD, Exac, G1K), impact, consequence, mode of inheritance in the case of Trio VCF files, etc. In addition, Variant Browser is integrated with the Seven Bridges Genome Browser and allows immediate visualization of the raw sequence reads for any given position.
During the BETA stage, you are able to interpret demo VCF files which are readily available inside the app. In case you want to interpret your own VCF files please contact us at [email protected].
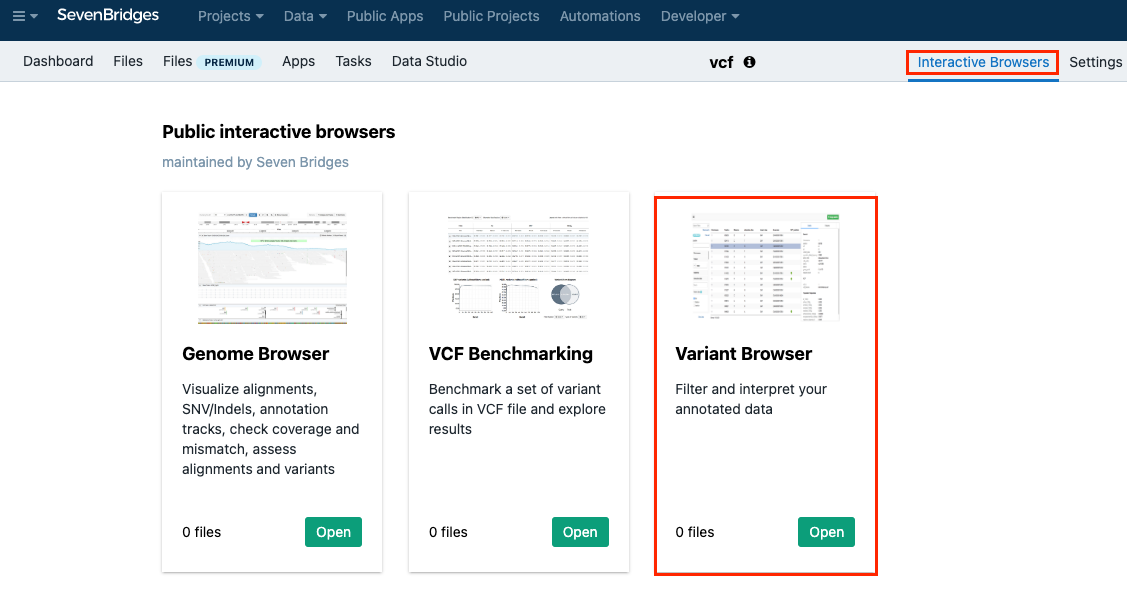
Access the Variant Browser
- Navigate to your project.
- Click Interactive Browsers in the upper right corner.
- Click Open within the Variant Browser section.
Interpret an annotated VCF file
To interpret a VCF file, click on any of the files in the Demo VCF files section. The page for filtering and interpreting VCF files is displayed.
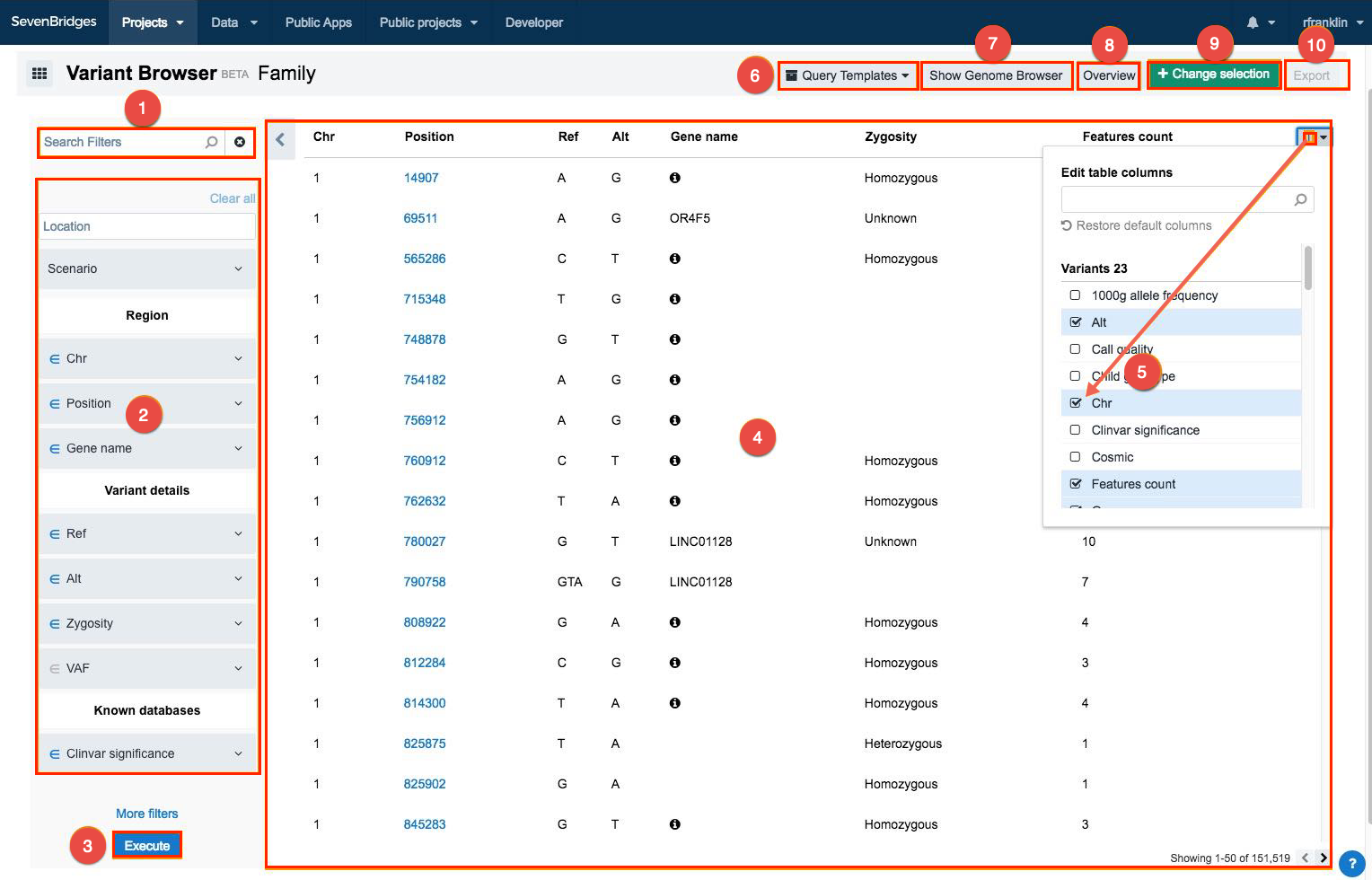
The following options are available:
- Search for a filter.
- Click on a filter to set the filtering criteria:
- You can define multiple values for a filter by pressing Enter after each one.
- Some filters have a "has value" option which you can use to filter all variants that have any kind of value defined.
- Use this
 symbol to decide if you want the specified value to be included or excluded from the results. For example, enter "NOC2L" for the Gene name filter. If you don't click the
symbol to decide if you want the specified value to be included or excluded from the results. For example, enter "NOC2L" for the Gene name filter. If you don't click the  symbol, the results will only show this gene. However, if you click the symbol, all genes but this one will be shown.
symbol, the results will only show this gene. However, if you click the symbol, all genes but this one will be shown. - Click More filters to display all available filters.
- Apply the selected filters.
- View results.
- Click this icon
 in the upper right corner to manage columns.
in the upper right corner to manage columns. - Query templates - click to manage the query templates:
- you can save your current query or open a previously saved template and continue from there
- when you save a template, you can use it across all of your projects; the templates that were saved by other users will only be available in the projects where they were originally created.
- Show Genome Browser - allows displaying variant positions in the Genome Browser; click to choose the related BAM file(s) and then click the position column for a variant to see its position in the Genome Browser.
- Overview - click to display general statistics for the currently applied filters (see below).
- Change selection - choose another input file.
- Export - this feature is not available during the BETA stage. Once it has been enabled, it will allow you to export results in CSV format and import them in another application.
Display general statistics
Once within the Variant Browser app, click Overview to display general statistics for the currently applied filters. The following information is available:
- Variant allele fraction
- Top mutated genes
- Ts/Tv Ratio
- Variants quality
- Variant types
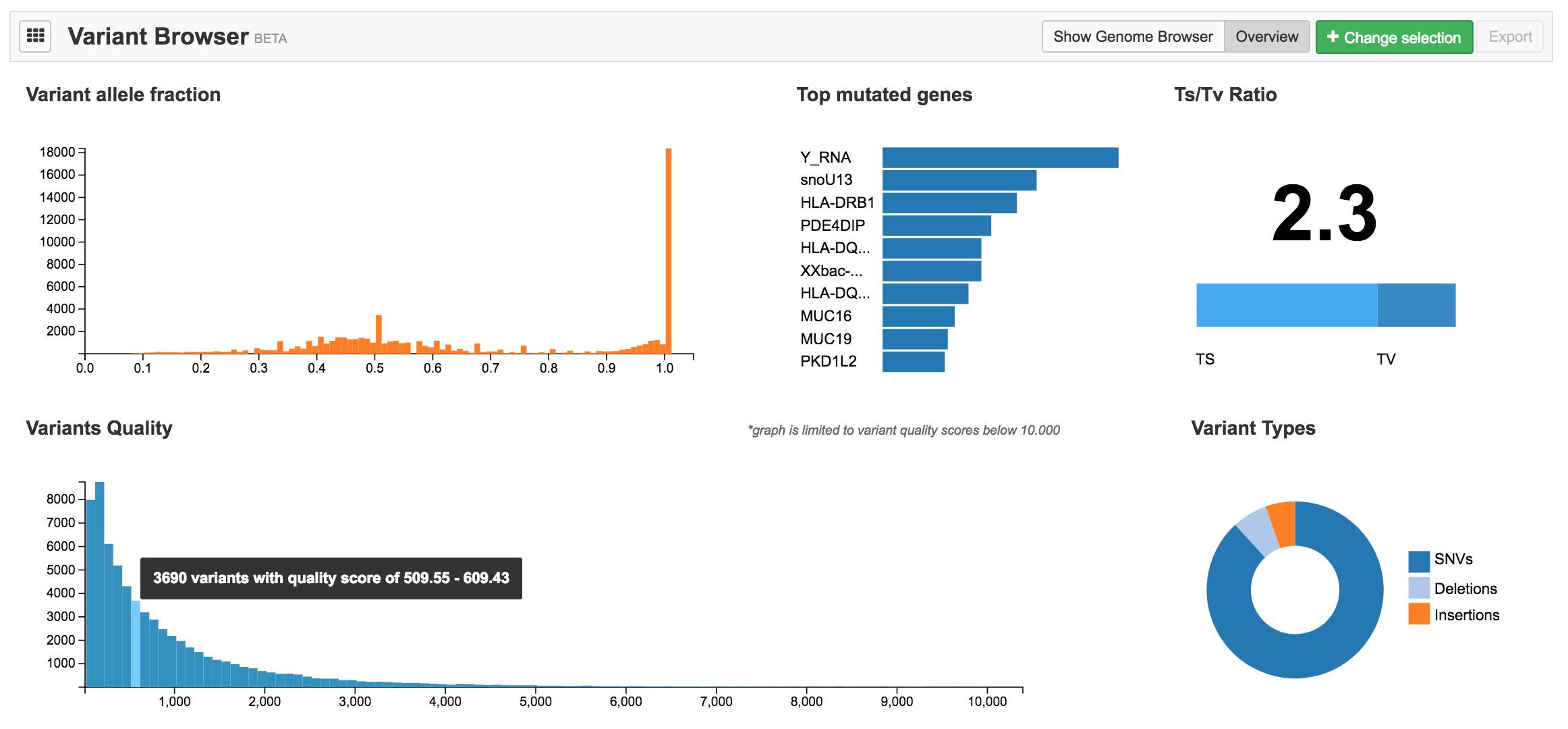
Hover your mouse over a diagram to display more detailed information.
An example of filtering a VCF file
- Click the NA12878.vep.variantbrowser_sqlite VCF file in the Demo VCF files section.
- Search for the GnomAD AF filter and click on it.
- Select <= and enter 0.01. This is done to filter out the common variants and find rare mutations which are potential causes of the disease.
- Next, search for the Call Quality filter, choose > and enter 80. This will result in showing only variants with value of QUAL field larger than 80.
- Then, search for the ClinVar significance filter and select Pathogenic.
- Next, search for Variant Category and enter coding:missense variant.
- Finally, search for Pathway and enter hemostasis.
- Click Execute.
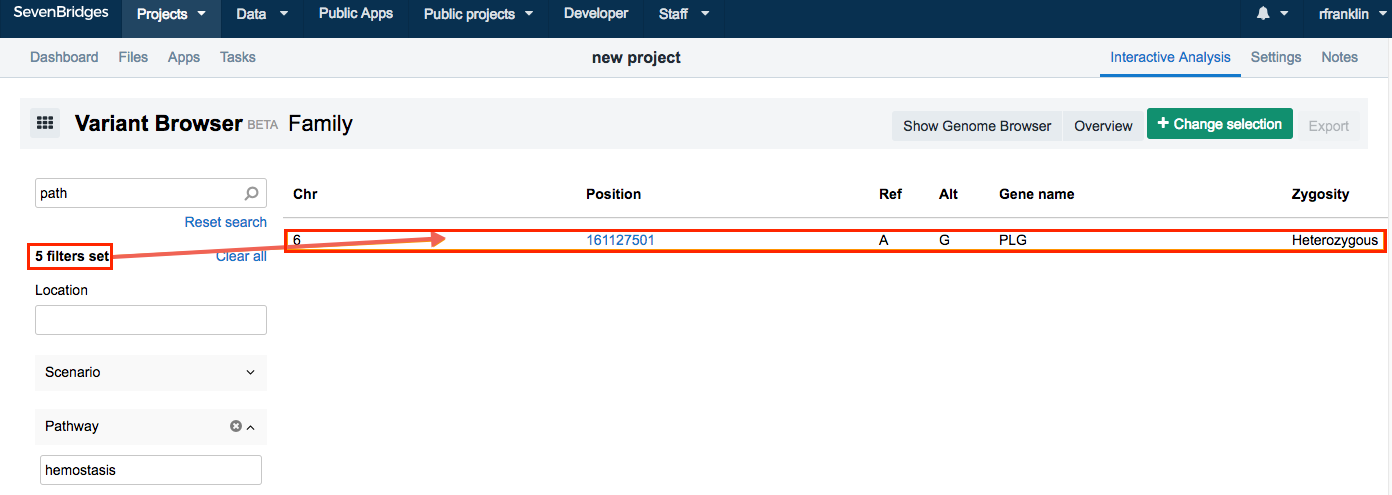
The four applied filters will filter out everything but one variant. Click on the variant and the pane with options Details, Features and Notes is displayed.
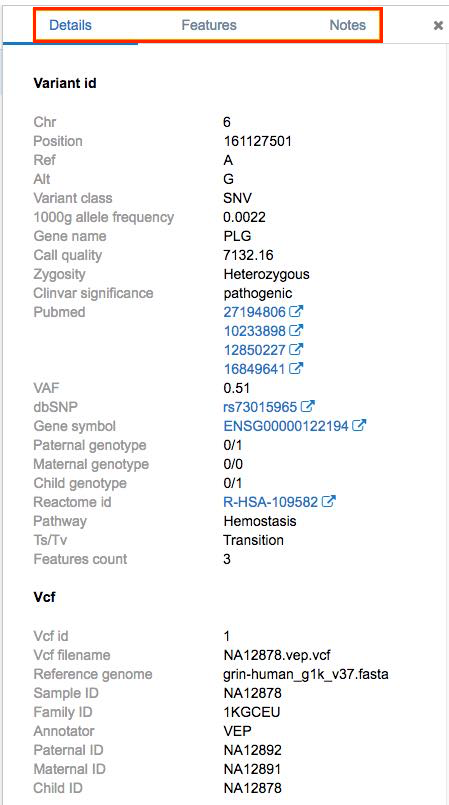
Variant Details
The Details tab gives you all available information about a variant, along with the links to external databases:
- dbSNP
- COSMIC
- PubMed
- Reactom database for pathways
- Ensemble database
- Uniprot/Swissprot
Variant Features
The Features tab lists all features (possible consequences) of the variant that match the applied filters. The number of features is also visible in the “Features Counts” column in the results page.
Variant notes
The Notes tab contains notes about the variant. All project members can use notes to exchange information or discuss ideas related to the variants. All members within a project can add notes or comment on an existing note.
Updated 3 months ago
