Overview
You can use the resources in the VCF Benchmarking Project to benchmark a set of variant calls in your VCF file and to interactively explore the results from a workflow execution (a "task").
This functionality should not be confused with VCF Benchmarking app which is used for analyzing and comparing benchmarking results obtained using this workflow.
Access the VCF Benchmarking project
Open the VCF Benchmarking Project by clicking on Public projects in the top navigation bar and then choose VCF Benchmarking.
The easiest way to benchmark your own set of variant calls is to:
- Copy the entire VCF Benchmarking Project to your own Seven Bridges account.
- Upload the VCF file you wish to benchmark to the Seven Bridges Platform.
- Add the VCF file to the VCF Benchmarking Project you copied in step 1.
- Modify the Draft task we have prepared in that project by entering your own input files and (optional) configuring the app settings.
- Run the task.
- View the results
The VCF Benchmarking Workflow (pipeline)
The VCF Benchmarking Project was prepared according to the recommendations of the Global Alliance for Genomics and Health (GA4GH) Benchmarking Team.
The data files contained in this project were produced by the GA4GH Benchmarking team and the Genome in a Bottle Consortium. The VCF Benchmarking Project contains the VCF Benchmarking Workflow (a "pipeline"), the tool you need to benchmark variant calls.
We've also packaged the necessary data files you need to run the workflow in this project.
The VCF Benchmarking Workflow closely follows the Benchmarking Pipeline Architecture defined by the GA4GH Benchmarking team. We implemented the VCF Benchmarking Workflow using the Common Workflow Language (CWL).
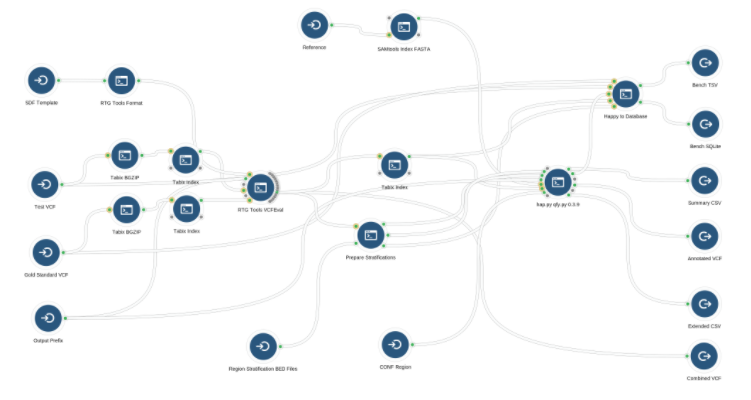
The VCF Benchmarking Workflow, as shown below, is built out of the following components:
- A comparison engine that produces a two-column VCF file compatible with the GA4GH specification. Our VCF Benchmarking Workflow uses the RTG Tools VCFEval tool as the comparison engine.
- The Prepare Stratification app which uses the bcftools view tool and a list of user specified character straficitions to produce a list of stratification BED files.
- The hap.py qfy.py tool which performs quantification using the two-column GA4GH VCF file produced by RTG Tool VCFEval.
- The Happy to Database app which writes the benchmarking results into a SQLite database file.
After it is successfully run, the VCF Benchmarking Workflow outputs the benchmarking results in an SQLite database file.
Stratifications
The VCF Benchmarking Workflow calculates benchmarking results by using four levels of stratifications to each consider a different subset of the variant calls:
- Variant type stratification: This categorizes variants by type. The current version of the VCF Benchmarking Workflow calculates the results for three categories: All, SNP, and Indel variants.
- Filtered variants stratification: This categorizes variants according to their filter status. Currently, two categories are used: (1) only the variants that pass all filters (‘.’ or ‘PASS’ in the VCF filter field), and (2) all of the variants in the VCF, regardless of filters.
- Region based stratification: This only considers variants in specific regions. A separate category is created for each of the input BED files. An additional category is calculated without applying any of the BED files.
- Character based stratification: This categorizes the variants by certain characters available as annotations in the two-column GA4GH VCF file. The current workflow performs stratifications using expressions in a format supported by the bcftools view tool. For example, you can stratify variants using the QQ field which is the variant quality used for creating a ROC plot. The expressions using different thresholds would be: “QQ <= 100”, “100 < QQ && QQ <= 200”, “200 < QQ && QQ <= 300”, etc.
You can set any combination of stratification parameters. The VCF Benchmarking Workflow produces results for the stratifications you select.
Learn more about benchmark stratifications from the GA4GH Benchmarking Team.
Precision/Recall Curves
The VCF benchmarking results may also include information for plotting Precision/Recall Curves. In order to plot the curves, tools need to be told the covariate(s) from the VCF against which to calculate the values. By default, the QUAL field (phred-scaled quality score) is used. This can be changed in the settings of the RTG Tools VCFEval and hap.py qfy.py tools in the VCF Benchmarking Workflow.
Comparison engine
The VCF Benchmarking Workflow uses the RTG Tools VCFEval tool as the comparison engine by default, but any tool that produces a two-column VCF file compatible with the GA4GH specification may be used. Changing the comparison engine is fairly straightforward, and requires changing the CWL App node in the workflow editor. In addition, you should make sure that all of the annotations used by the workflow are written out by the comparison engine used. By default, the workflow requires the the QUAL field (phred-scaled quality score).
Inputs
The VCF Benchmarking Workflow requires several inputs to work properly, as shown below. Inputs are represented by circular nodes with an arrow pointing into a semi-circle  .
.
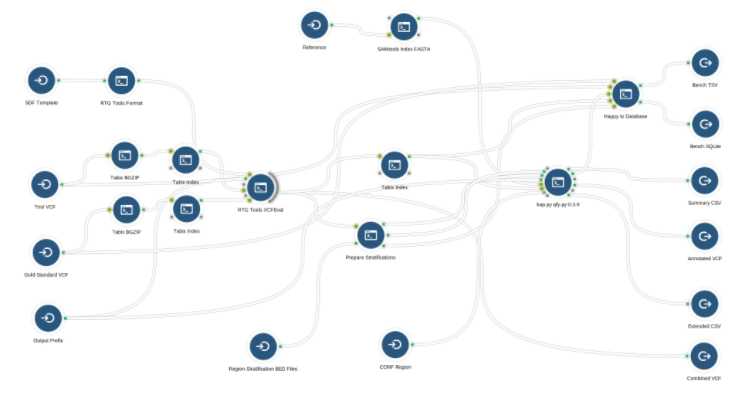
Inputs for the workflow are as follows:
File | Description |
|---|---|
Test VCF required | This file contains the variant calls you wish to benchmark. The Test VCF needs to be produced using a reference file which is compatible with the reference file used to produce the Gold Standard VCF. |
Gold Standard VCF | This file contains a set of variant calls that are the standard against which the Test VCF is benchmarked. Currently there are Gold Standard VCF files for 5 Genome in a Bottle samples (HG001-HG005) for both reference genome GRCh37 and GRCh38. |
Reference FASTA | This FASTA file contains the reference genome used to produce both the Test VCF and Gold Standard VCF files. |
SDF Template File | A pre-formatted reference dataset used by the RTG Tools VCFEvaltool. If no SDF file is available, the same reference FASTA file used to produce the input VCF files can be passed in. |
Region Stratification BED Files | These BED files are used to provide regional stratifications by considering variants in specified regions. None, one, or multiple BED files may be provided. The workflow always produces one set of results for the entire genome as well as any regionally stratified results. |
CONF BED File | A BED file containing confident regions. If specified, all regions outside this BED file will be labeled as unknown. |
Output Prefix | The output prefix for all output files produced by the workflow. |
Character Expression | (optional) Character stratifications performed using expressions in a format supported by the bcftools view tool. Variant stratification can be done using the QQ field which is the variant quality used for creating a ROC plot. E.g. “QQ <= 100”, “100 < QQ && QQ <= 200”, “200 < QQ && QQ <= 300”, etc.: A setting of the RTG Tools VCFEval tool. Allow alleles to overlap where bases of either allele are same-as-ref. Default is to only allow VCF anchor base overlap. |
Sample | A setting of the RTG Tools VCFEval tool. If using multisample VCF files, set the name of the sample to select. Use the form baseline_sample, calls_sample to select one column from the Gold Standard VCF and one column from the Test VCF, respectively. |
Squash Ploidy | A setting of the RTG Tools VCFEval tool. For a diploid genotype, both alleles must match in order to be considered correct by RTG Tools VCFEval. To identify variants which may not have a diploid match but which share a common allele, use this option. |
Results
The VCF Benchmarking Workflow outputs the benchmarking results in a SQLite database file. This file will be produced in the project, and shown under the Files tab, after you have completed the VCF Benchmarking Workflow. The calculated values reflect the GA4GH Benchmarking Team’s recommendations Comparison Method #3. The SQLite database contains a separate entry for each of the calculate stratification combinations.
The database schema is shown below.
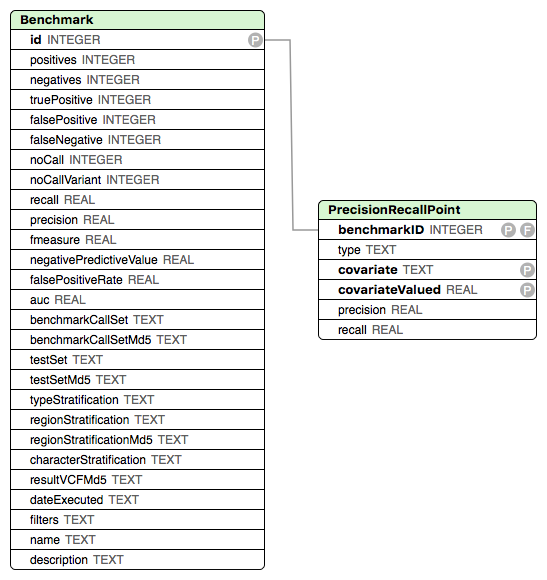
Field definitions in Benchmarking Result File
Term | Description |
|---|---|
ID | The ID of the benchmark |
Positives (P) | The number of variants in the Truth VCF file |
Negatives (N) | The number of hom-REF bases in the Truth VCF file; currently not calculated by the VCF benchmarking workflow |
True Positive (TP) | The number of variants in the Query VCF file that match the Truth VCF file |
False Positive (FP) | The number of variants in the Query VCF file that do not match the Truth VCF file |
False Negative (FN) | The number of variants in the Truth VCF file that do not match the Query VCF file |
noCall | The number of no-call sites in the Truth VCF file; currently not calculated by the VCF benchmarking workflow |
noCallVariant | The number of no-call sites in the Query VCF file; currently not calculated the VCF benchmarking workflow |
Precision | True Positive / (True Positive + False Positive) |
Recall | True Positive / (True Positive + False Negative) |
F-measure | 2_Precision_Recall/(Precision+Recall) |
Negative Predictive Value | False Positive / (False Positive +False Negative); currently not calculated by the VCF benchmarking workflow |
False Positive Rate | False Positive / Negatives; currently not calculated by the VCF benchmarking workflow |
AUC | Area under the ROC curve; currently not calculated by the VCF benchmarking workflow |
BenchmarkCallSet: | The name of the call set that was used as the Truth VCF. By default, the VCF benchmarking workflow uses the name of Truth VCF file |
BenchmarkCallSetMd5 | The MD5 checksum (a cryptographic hash value) for the Truth VCF file |
TestCallSet | The name set of variants that was benchmarked. By default, the VCF benchmarking workflow uses the name of the Query VCF file |
TestCallSetMd5 | The md5 checksum of the Query VCF file |
TypeStratification | Type of variants taken into the account when calculating the results (All, SNP, Indel, etc.) |
RegionStratification | Region from which the variants were taken into the account when calculating the results. Provided as a BED file to the pipeline; the pipeline uses the name of the BED file for this value. |
RegionStratificationMd5 | The md5 checksum of the BED file used as region stratification |
CharacterStratification | The character-based stratification used to calculate the results |
ResultVcfMd5 | The md5 checksum of the intermediate VCF file produced by the comparator |
dateExecuted | The date the benchmark was executed |
Filters | Filter based stratification that was applied (filters that were taken into the account when calculating the results) |
Name | The name of the benchmark |
Description | A description of the benchmark |
PrecisionRecallPoint table
| BenchmarkID | The ID of the benchmark this Precision/Recall point belongs to |
| Type | The variant type based stratification |
| Covariate | The covariate against which the Precision/Recall curve was calculated |
| CovariateValue | The value of the covariate at this Precision/Recall point |
| Precision | True Positive / (True Positive + False Positive) |
| Recall | True Positive / (True Positive + False Negative) |
Run a VCF benchmarking analysis
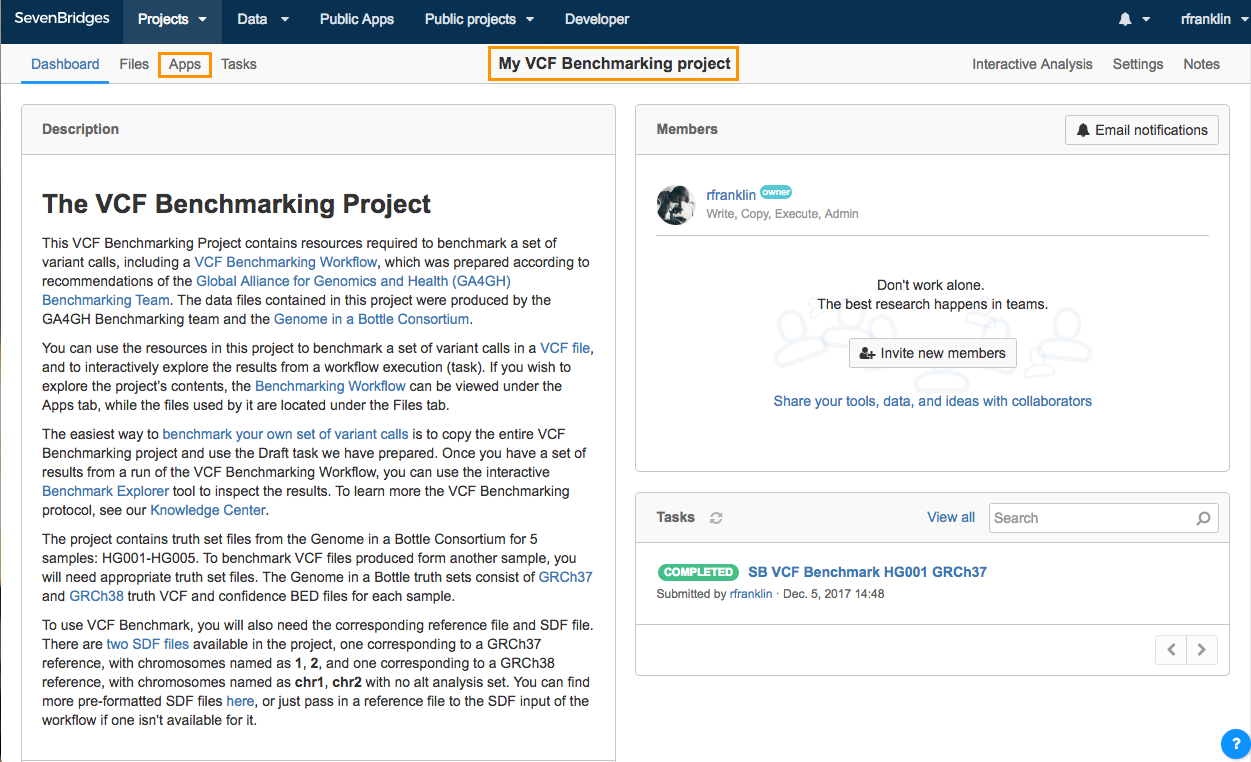
To benchmark your VCF file, you need to first make your own copy of the public VCF Benchmarking Project. Then, upload your VCF file to the Platform and add it to your copy of the VCF Benchmarking Project. Then, set up your task using using the existing Draft task under the Tasks tab.
Copy the VCF Benchmarking Project
- Click Public projects on the top navigation bar.
- Select VCF Benchmarking.
You will be taken to the VCF Benchmarking Project dashboard. - Click the info icon next to the project title. Project information box is displayed.
- In the bottom-right part of the project information box, click Copy project.
- Name your copy of the project, select a billing group and modify execution settings if needed.
- Click Copy
Once the project has been copied, you will be taken to your project which contains a copy of the VCF Benchmarking Workflow as well as necessary files.
Set up a new task
To set up a new task, navigate to the Tasks tab in your VCF Benchmarking Project. We have prepared 10 Draft Tasks for you to start your analysis. There is a draft task for each combination of a Genome in a Bottle sample (HG001-HG005) and supported reference type (GRCh37 and GRCh38). There is also a completed task for sample HG001 for the reference type GRCh37.
To start setting up your task, click the name of the desired draft task. In the example below, the task is named SB VCF Benchmark HG001 GRCh37.
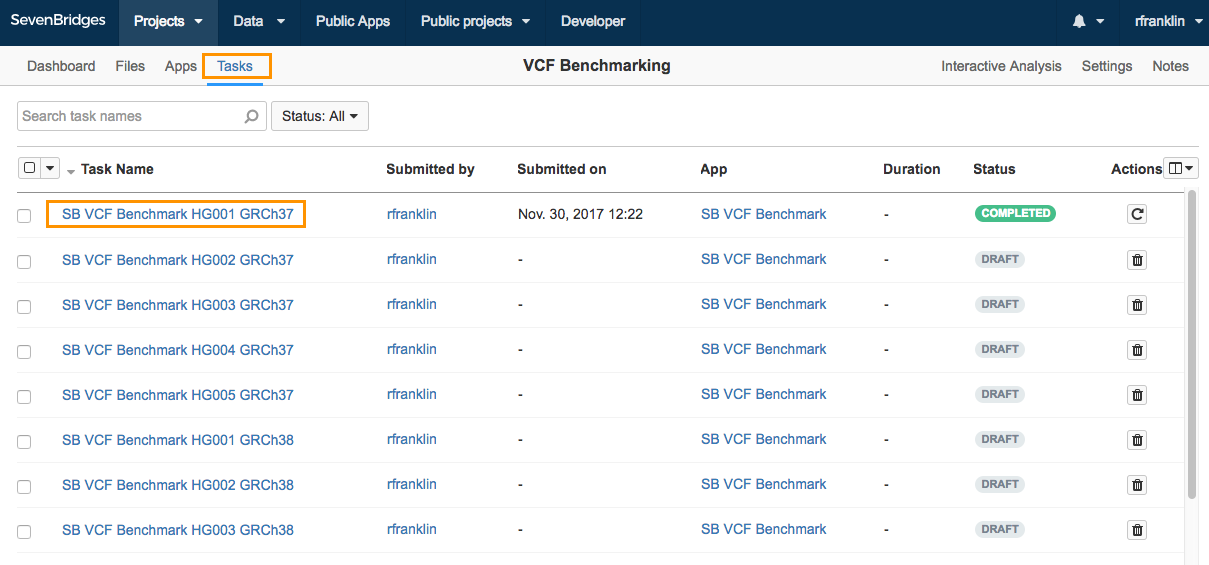
On the Draft task page, you can specify the input files and define app parameters. Here, you can see that some of the required input files (the reference file, stratification BED files, Gold Standard VCF and SDF template) have already been added. If the Test VCF file that you are benchmarking is for the HG001 sample, and is produced with the GRCh37 version of the reference genome, the only input file you need to provide is the Test VCF.
In that case, all you need to do is set your VCF file as the Test VCF input.
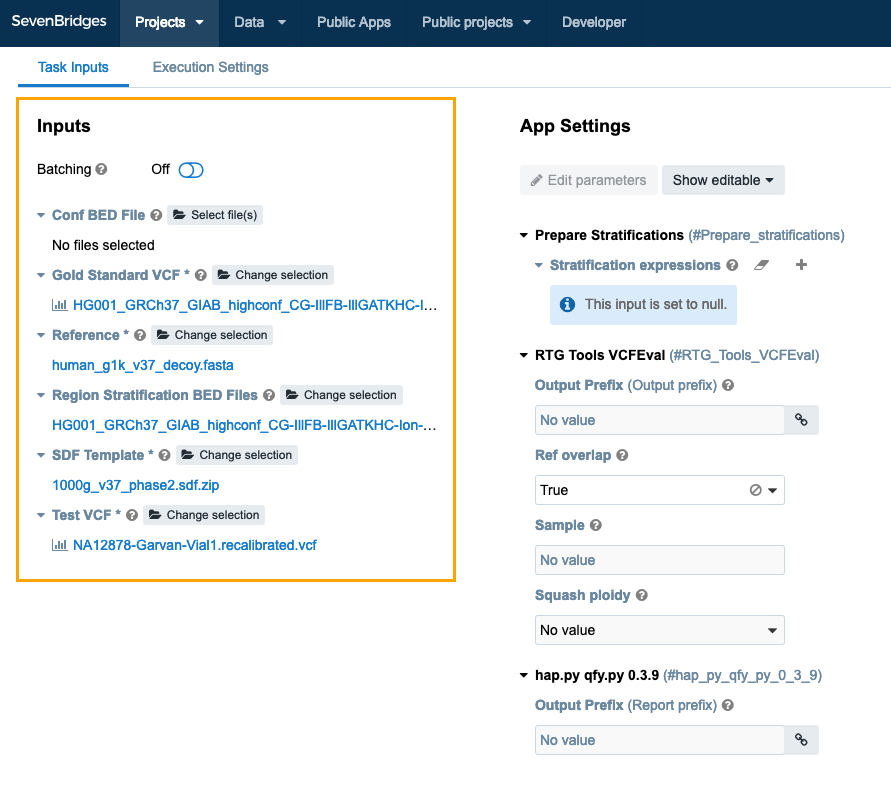
Select Input files
The VCF Benchmarking Workflow requires several inputs to work properly. The default input files provided with the VCF Benchmarking Workflow are intended for benchmarking against the Genome in a Bottle NA12878 sample, processed with the GRCh37 reference genome. If the Test VCF file that you are benchmarking is for the NA12878 sample, and is produced with the GRCh37 version of the reference genome, the only input file you need to provide is the Test VCF. This is the VCF file you wish to benchmark.
To select the Test VCF file for your task, click Select file(s) (below Test VCF). You'll be taken to file picker to select the desired file. Note that you will need to have added your Test VCF in your project. To easily locate a file, you can search by its name or filter by file type (VCF). To choose the file, tick the checkbox next to it and click Select. This takes you to the Set Input Data page with the newly selected file.
The pre-populated files should only be used if the your Test VCF file is for the NA12878 sample, and produced with the GRCh37 version of the reference genome. Learn more about running the VCF Benchmarking Workflow on a custom dataset.
Define App Settings (optional)
The App Settings column on the Draft Task page allows you to enter optional information to customize your analysis.
- Output Prefix: The output prefix for all output files produced by the workflow.
- Character Expressions: Character stratifications performed using expressions in a format supported by the bcftools view tool. Variant stratification can be done using the QQ field which is the variant quality used for creating a ROC plot. E.g. “QQ <= 100”, “100 < QQ && QQ <= 200”, “200 < QQ && QQ <= 300”, etc.
- Ref Overlap: A setting of the RTG Tools VCFEval tool. Allow alleles to overlap where bases of either allele are same-as-ref. Default is to only allow VCF anchor base overlap.
- Sample: A setting of the RTG Tools VCFEval tool. If using multisample VCF files, set the name of the sample to select. Use the form baseline_sample, calls_sample to select one column from the Gold Standard VCF and one column from the Test VCF, respectively.
- Squash Ploidy: A setting of the RTG Tools VCFEval tool. For a diploid genotype, both alleles must match in order to be considered correct by RTG Tools VCFEval. To identify variants which may not have a diploid match but which share a common allele, use this option.
Run Your Analysis
Once you select all the necessary input files on the Draft Task page, click Run to start your analysis. Once the task has finished, it will output an SQLite database file. You can download this file to view results.
View Results
Once the task finishes, it will output an SQLite database file.
To view the SQLite database file using the Benchmark Exploration tool:
- Navigate to the Files tab in your project.
- Click the file with SQLite extension.
To download the SQLite database file:
- Navigate to the Files tab in your project.
- Tick the checkbox next to the file with sqlite extension.
- Click Download.
Run benchmarks on custom datasets
Benchmark against samples other than HG001
In order to benchmark a set of variant calls against samples other than HG001, you need a Truth VCF that acts as the Gold Standard VCF against which to benchmark your VCF file. In the public VCF Benchmarking project, we prepared Gold Standard VCF files for 5 Genome in a Bottle samples, HG001-HG005, for both reference types GRCh37 and GRCh38.
There is a prepared Draft Task in the Tasks page for each combination of a sample and reference type.
To benchmark against different samples and/or reference types, you will need to change the inputs of workflow accordingly.
If this Gold Standard VCF, along with the Test VCF, is produced using the GRCh37 reference genome, all that you need to do is replace the Gold Standard VCF on the draft task page when starting a task. The draft task page will be pre-populated with the GRCh37 reference genome.
To pick a new Gold Standard VCF:
- Navigate to the draft task page.
- Click Pick files below Gold Standard VCF.
- Select the desired Gold Standard VCF from the file picker by ticking the checkbox next to its name.
- Click Select.
Benchmark against different samples and/or reference types
In order to benchmark a Test VCF that isn’t one of the 5 Genome in a Bottle samples, you need to provide the following inputs:
- The Gold Standard VCF for the desired sample
- A list of BED files to be used for stratifications (optional)
In order to benchmark a Test VCF that is not produced with the GRCh37 reference genome, you need to provide the following input files:
- The Gold Standard VCF for the desired version of the reference genome
- The FASTA file with the desired version of the reference genome
- The SDF template or FASTA file with the desired version of the reference genome
- A list of BED files to be used for stratifications (optional)
Edit the VCF Benchmarking Workflow
Modify Variant Stratification
The VCF Benchmarking Workflow is flexible in terms of the stratifications that can be processed. The Benchmark Exploration tool also offers a dynamic view of the specified stratifications in the output.
Region based stratification is the easiest to modify. This can be done by providing additional or different stratification BED files. In the output SQLite database file, the Region Stratification Name field will reflect the BED file name. Please ensure that the any provided BED file is compatible with the reference FASTA file and query and test VCF files.
Variant type stratification by default produces results for only SNPs, only INDELs, or both of these types bundled together.
Filter stratification specifies how to treat variants that have a non-passing filter value. “All” means apply all filters, i.e. variants with non-passing filter values are excluded. “None” means “ignore all filters”, i.e. variants with non-passing filter values are included in the calculation. By default, results are produced for both of these options.
The parameter which controls the character stratification is a list of selection expressions which are passed to the bcftools view tool to create subsets of variants. Each expression is taken as a separate stratification category. You may add or remove expressions, but make sure that all of the necessary fields are present in the two-column VCF file with the benchmarking results.
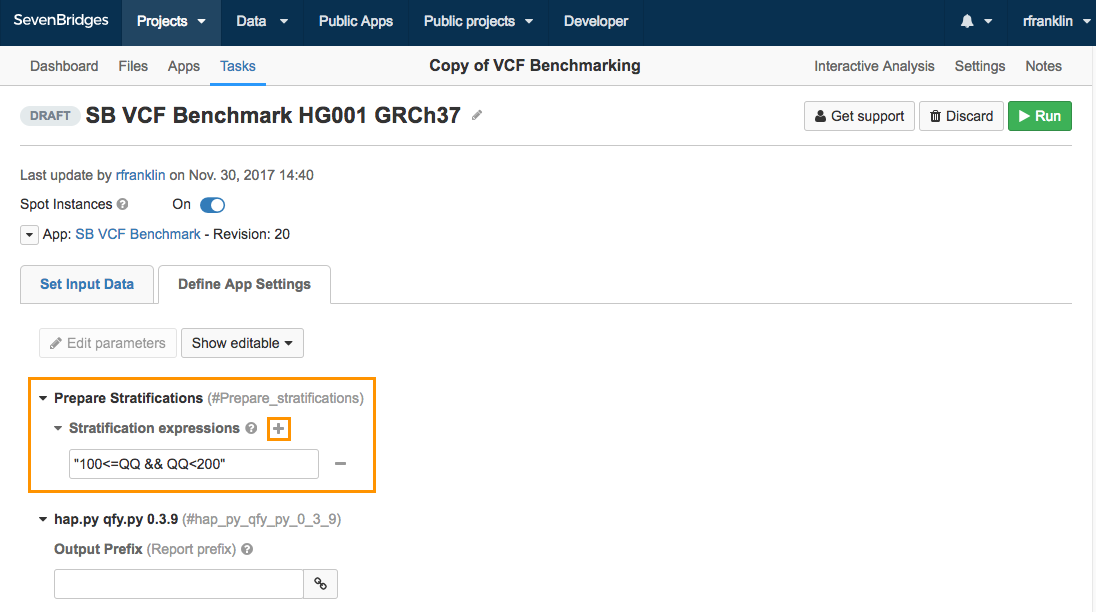
To modify a Character stratification parameter:
- Navigate to the draft task page.
- Go to the Define App Settings tab.
- Click the plus icon next to Stratification expressions.
- Add a new expression into the empty field.
Updated 3 months ago
