QuickStart
Prerequisites
Reach out to us to test drive the Platform.
Procedure
We'll start by creating a project and populating it with FASTQ files. Then, we'll use one of the Seven Bridges whole exome analysis workflows to carry out the analysis. Finally, we'll examine our results.
Create a project
The first step to running an analysis on the Seven Bridges Platform is to create a project. To do this, click Create a project under the Projects tab in the top navigation bar.
This will open a new window where you can name your project and select a billing group. When you're finished, click Create.
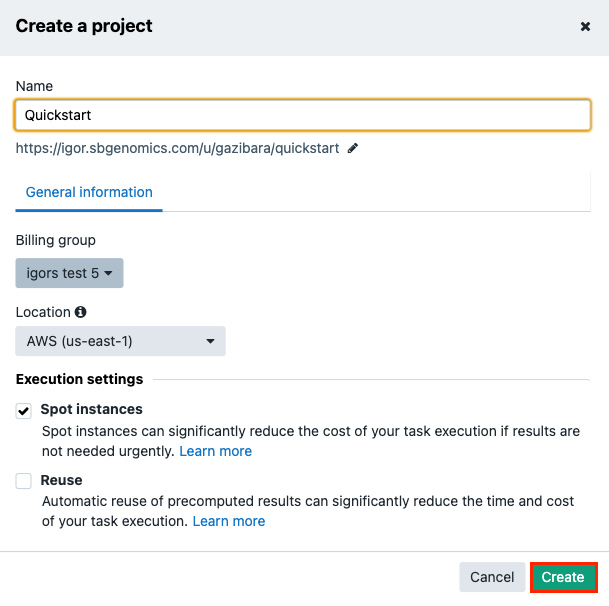
Once you create a project, you'll be taken to its Project Dashboard. This page contains all the information about your project, including its files, apps (tools and workflows), tasks (workflow executions), and project members.
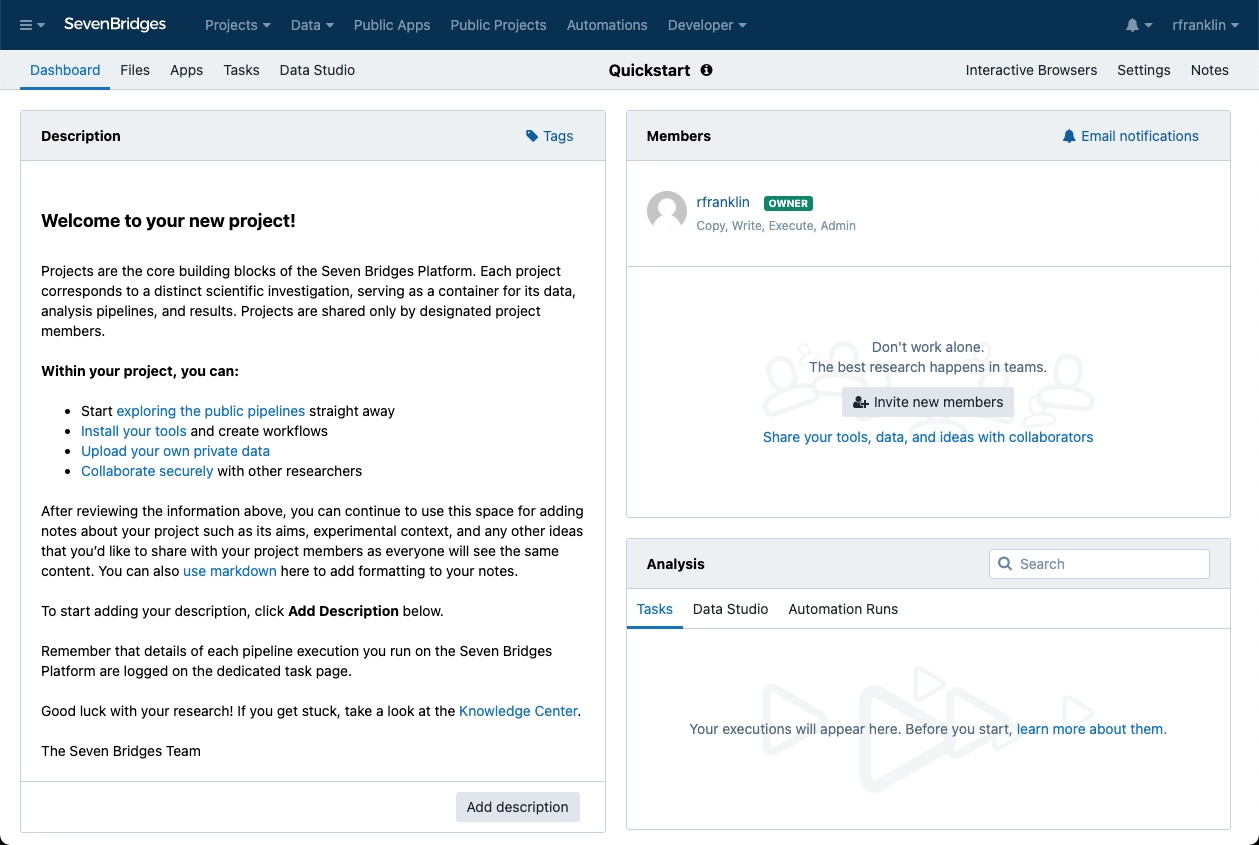
Add FASTQ files to your project
The next step is to add the FASTQ files to your project. The other reference files needed for the analysis will be suggested when you set up the workflow.
To find the FASTQ files necessary for the analysis, click the Files tab on your project dashboard and then Add files > Public Files.
For this analysis, we want to use two paired-end files that contain whole exome sequencing data called C835.HCC1143.2.converted.pe_1.fastq and C835.HCC1143.2.converted.pe_2.fastq.
Enter "C835.HCC1143.2.converted.pe_" into the search box to find the files.
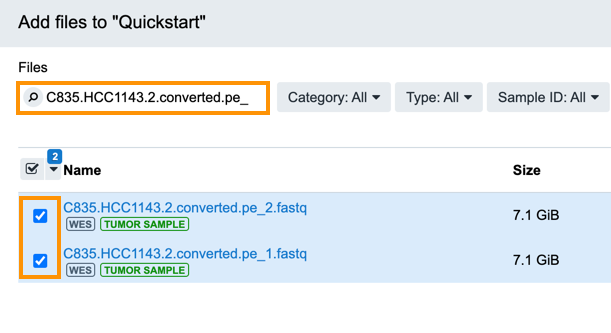
Select both files and click Copy to Project and confirm it.
Enter file metadata
It is important to annotate your files with Metadata on the Seven Bridges Platform when you perform an analysis on the Platform so that bioinformatics tools processing files in parallel can group files with identical metadata value(s) in specified fields.
File metadata includes information about the File (e.g. experimental strategy and library ID), Sample (e.g. sample ID), and General (e.g. investigation and species) . For more information on the metadata fields used on the Platform, please see the documentation on file metadata.
Click the Files tab on your project dashboard to see all the files in the project. Currently our project, QuickStart, only contains the two files that we've just added.
Select both of the files and click Edit Metadata. This will open a pop-up window with inputs for the different metadata fields. Notice the empty field for Platform unit ID. This needs to be set to run the task. Enter 1 in this field, and click Save.
This metadata will inform tools that these files come from the same sample, were produced by the same library, and have been sequenced on the same lane.
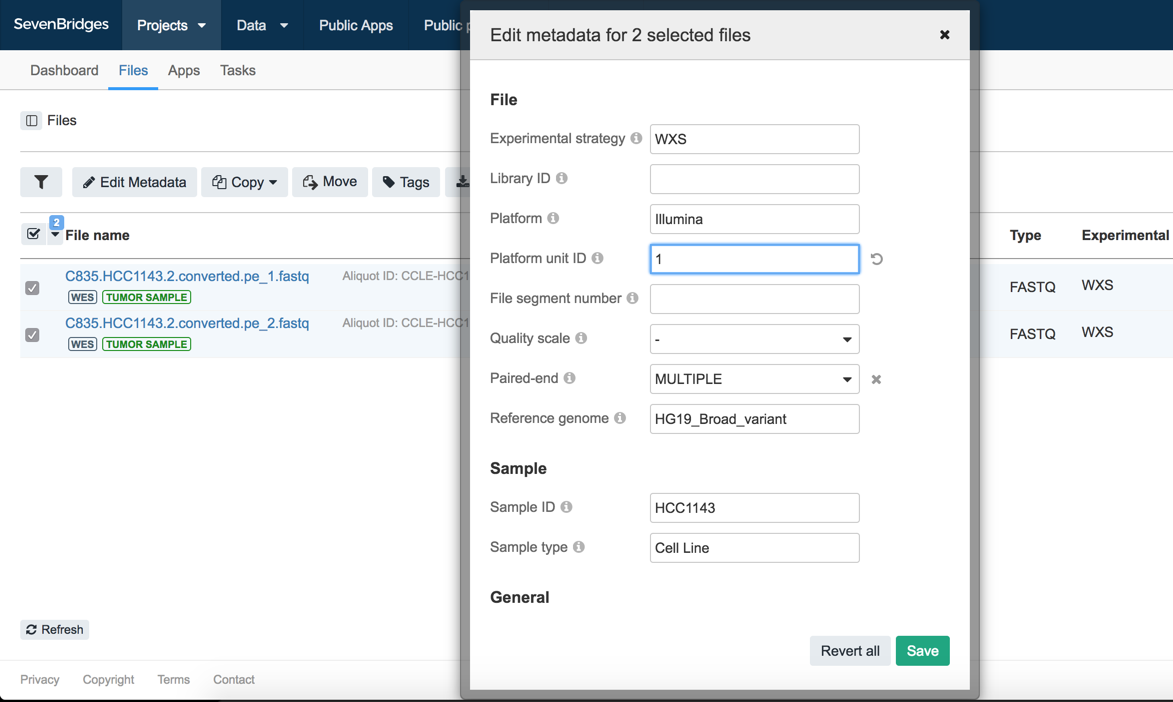
Each file used in an analysis on the Seven Bridges Platform must have their own metadata values. For more information, see the metadata documentation on grouping and distinguishing files by metadata.
In the example here, note that while we have set the same Library ID, Platform unit ID, and Platform values for the two FASTQ files, those two files come with different Paired-end values ('1' and '2') by default.
Select a public workflow
The next step is selecting a public workflow for running the analysis. We'll use the workflow, Whole Exome Analysis - BWA + GATK 4.0 (with Metrics), which is based on the free version of the GATK tool developed by the Broad Institute.
This workflow is one of Seven Bridges' many open source workflows available to all users on the Platform. These workflows have been tested to run efficiently in the cloud environment by the Seven Bridges bioinformatics team.
To select a public workflow for use in your project, navigate to Apps tab on your project dashboard and click Add App.
To add the Whole Exome Sequencing workflow:
- Type 'whole exome' into the search box. Whole Exome Sequencing - BWA + GATK 4.0 (with Metrics) will be displayed in the search results.
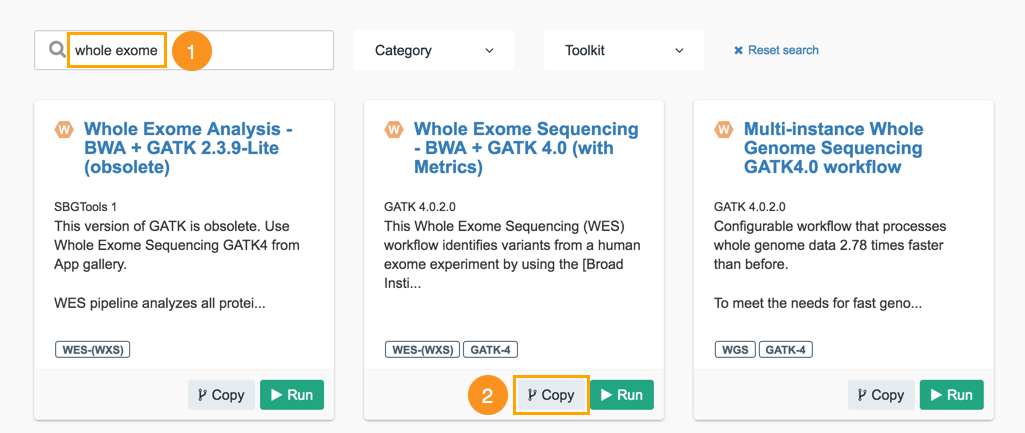
- Next, click Copy below the workflow.
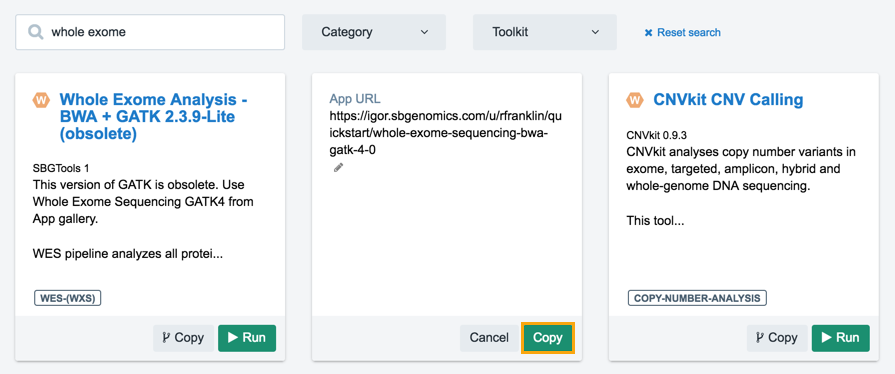
- Click Copy and the workflow will be added to your project.
To go back to the project dashboard, close the app browser window.
Edit the selected workflow
In many cases, you might want to tweak a workflow to work better with your dataset. This can be done easily using the workflow editor. To edit your workflow in your project, navigate to the Apps tab, click the ellipsis icon next to Whole Exome Sequencing - BWA + GATK 4.0 (with Metrics) and select Edit.
This opens the workflow editor containing a graphical representation of the workflow where each tool, input, and reference file is represented as a node.
To the right of the workflow diagram, the panel labeled APPS displays a list of all the apps available in your projects (MyApps) or among PublicApps.
The PARAMS panel describes the parameters of the tools used in this workflow and allows you to make quick edits.
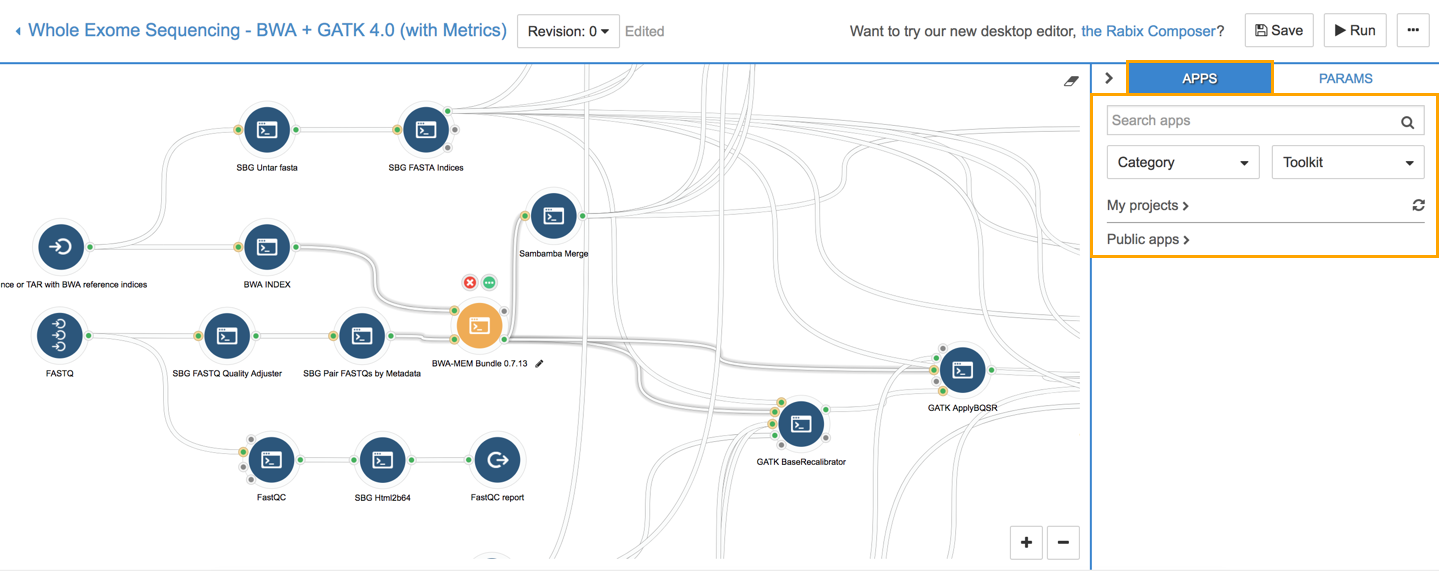
In the workflow editor, click the BWA-MEM Bundle 0.7.13 node (see the screenshot below). This opens the PARAMS tab, which displays the parameters of BWA-MEM Bundle sorted into Input/Output options, Scoring options, Execution, etc. Select the use_soft_clipping parameter.
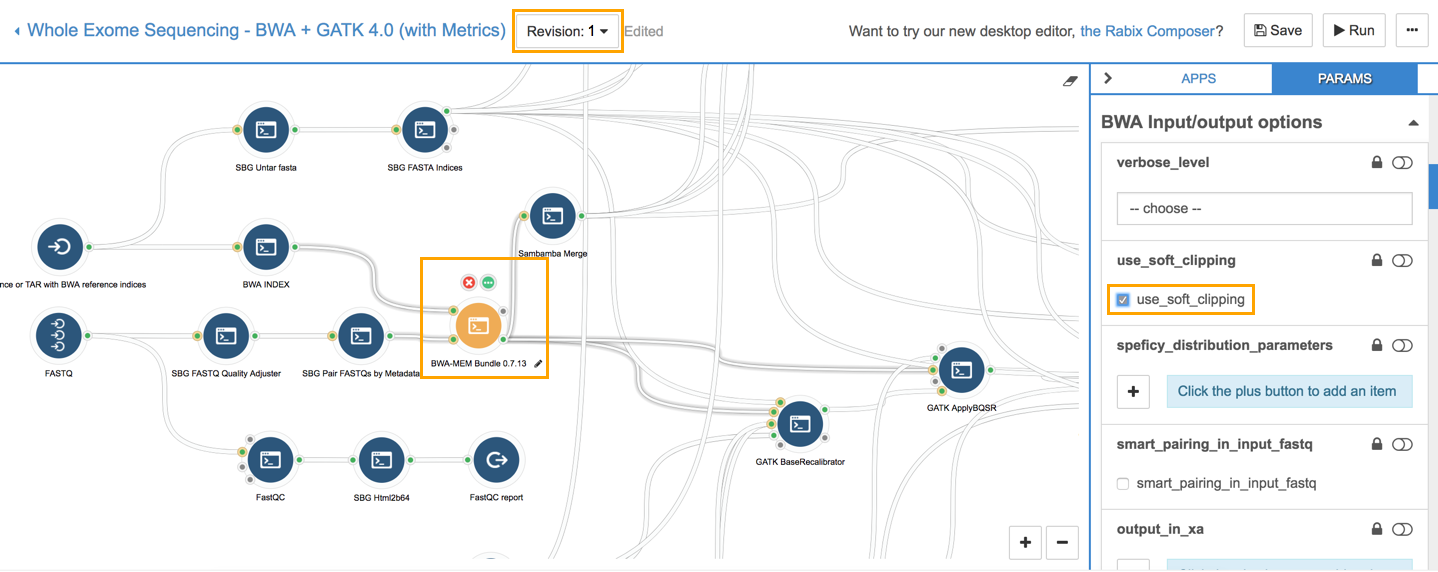
This will soft clip the supplementary alignments. To save this change as a new revision of the workflow, click Save. Note that clicking Save changes the version number from 0 to 1. This function allows you to keep track of all your workflow edits.
Run the analysis
Now that the workflow is ready, it's time to run the analysis. We'll click Run, in the upper right corner of the Tool Editor. The DRAFT task page will open with a pop-up window containing suggested files for this workflow.
For all public workflows on the Seven Bridges Platform our team of bioinformaticians has chosen a set of recommended input files.
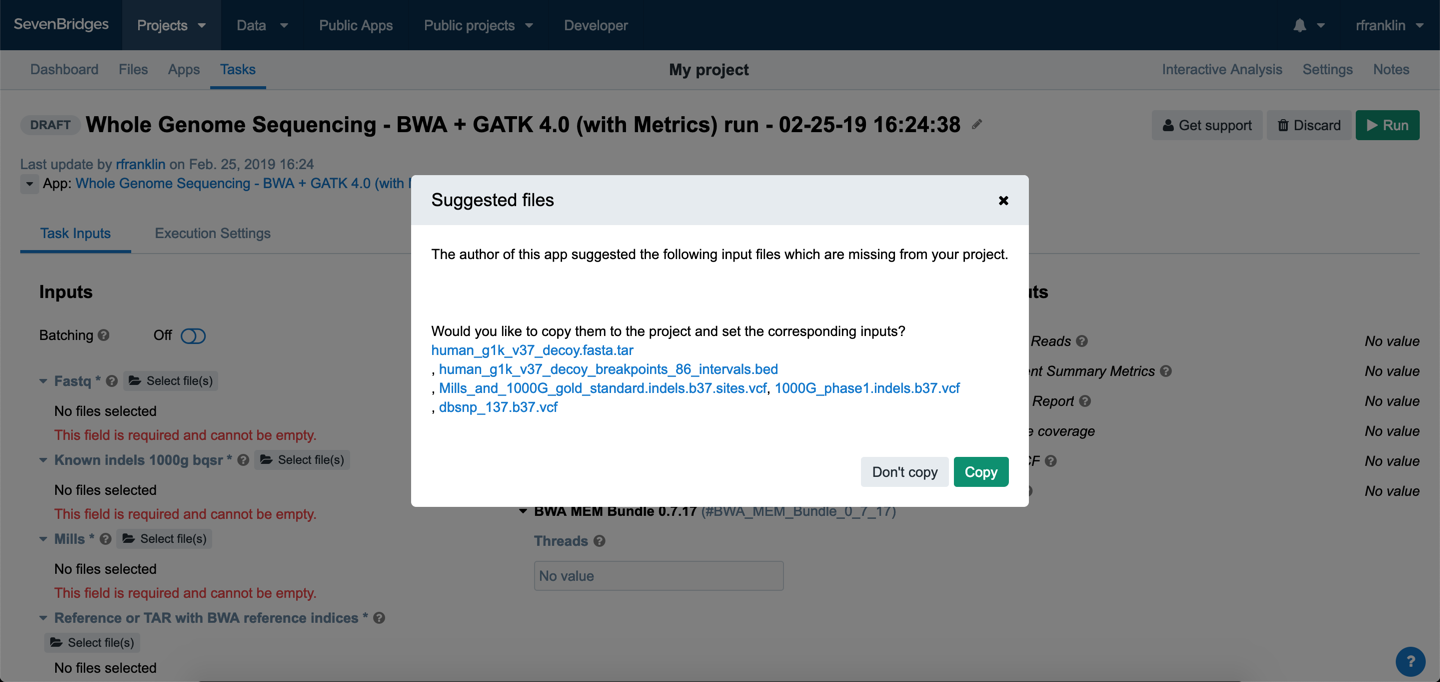
Click Copy and the suggested files will be copied to your project and automatically added to the matching input ports of your workflow. The files are mapped the following way.
Input port | Input files | File type |
|---|---|---|
Known_SNPs Known_Indels | dbsnp_137.b37.vcf Mills_and_1000G_gold_standard.indels.b37.sites.vcf 1000G_phase1.indels.b37.vcf |
|
Target_BED | exome_targets.b37.sorted.bed |
|
SnpEff_Database | GRCh37.75.zip |
|
FASTQ | C835.HCC1143.2.converted. |
|
Reference or TAR with BWA reference indices | human_g1k_v37_decoy.fasta |
|
On the DRAFT Task page you will see two tabs: Task Inputs and Execution Settings as shown below. The Task Inputs tab is where you can enter the input files and reference files for your workflow.
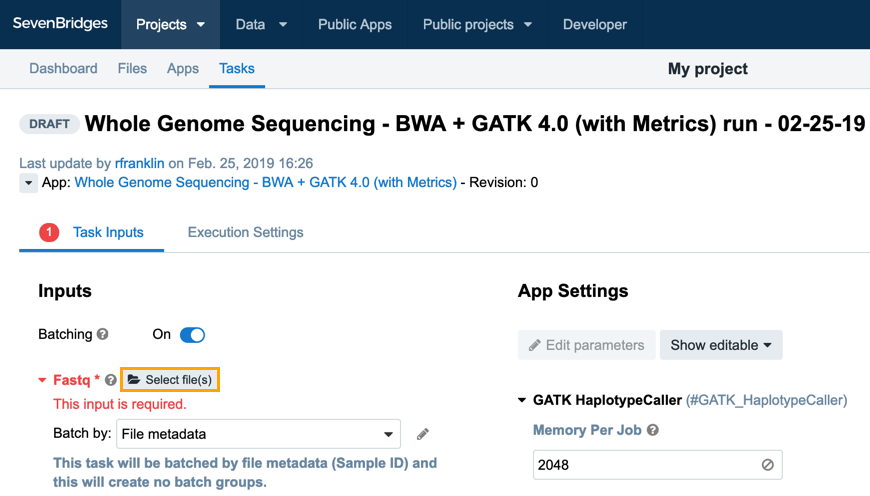
The only remaining files you need to select are FASTQ files. Click Select file(s) and choose these files:
- C835.HCC1143.2.converted.pe_1.fastq
- C835.HCC1143.2.converted.pe_2.fastq
The files will be batched by sample, meaning that files with the same Sample ID metadata field will be processed together in a separate task. In our case, the paired-end files we picked already had the Sample ID field set to the same value.
After adding the two FASTQ files, we can start this execution by clicking Run in the task page and then once again inside the popup to confirm.
When you start the task, a new page opens displaying the task's properties. To see all the tasks that have run or are running in this project, click Back to tasks in the upper left corner.
Here you can see the name of each task, the project member who started it, its initiation time, the execution workflow, its status, and available task actions.
The status will be a progress bar if the task is still running or a label notifying whether the task has completed, been aborted or failed.
Additional information, including how to check the status of the task or how to troubleshoot in case of the failed task, is available in the documentation on task statistics.
View the results of the data analysis
Once the task is completed, you'll be notified via email. The easiest way to access results is to go to the Tasks tab. This shows all the information related to this particular execution.
On the Tasks page, the column marked Outputs shows the results produced by the tools in the executed workflow. In our example task, take a look at summary_metrics report. Clicking on the file name opens the alignment metrics from the task.
At the bottom of the screen you can see the task's raw output.
The result of the data analysis is shown in the raw VCF file. The raw VCF contains all the variants detected by the workflow. To download it, just click on its filename. This will open a new page displaying the contents of the file and some information describing it. Then click Download in the upper right corner.
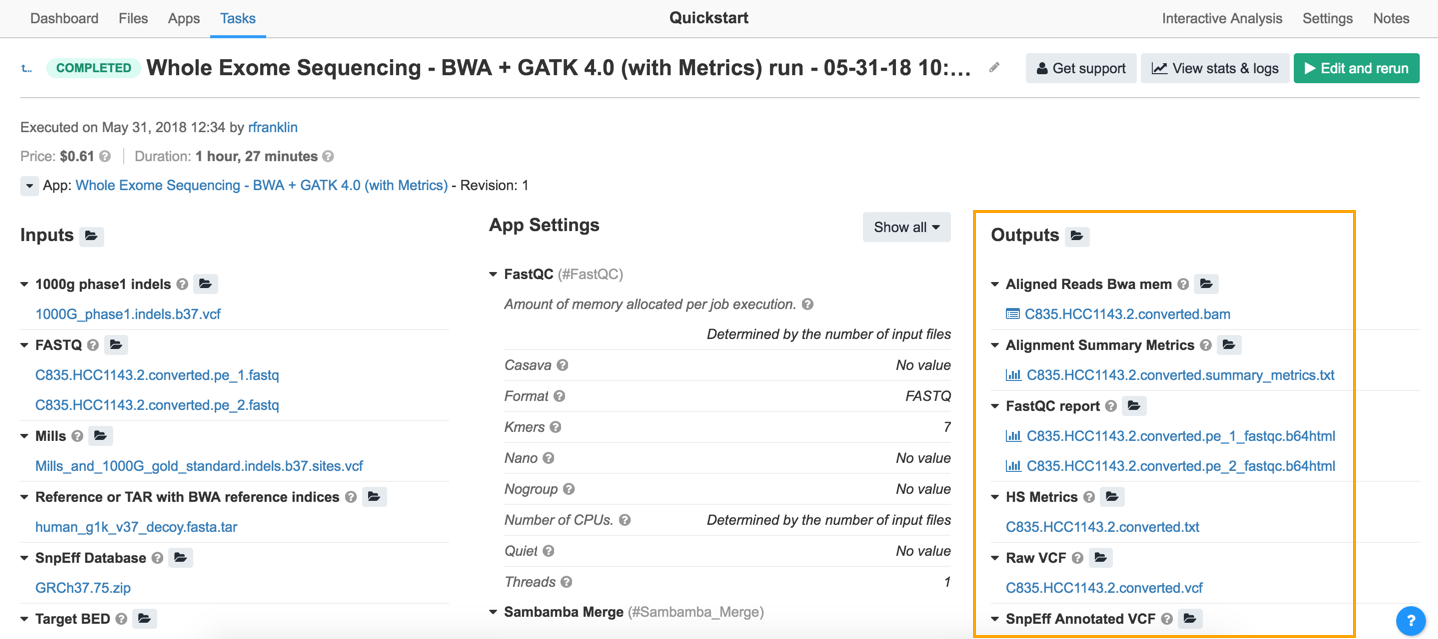
Note that the names of files outputted from a tool incorporate part of the tool's name. This makes it easier to find report files from a list of outputs.
That’s it! We've executed a data analysis and obtained some results. We encourage you to try this procedure for yourself before getting started on your own data analyses. You can also visit the rest of our Knowledge Center to learn more about the Seven Bridges Platform and bringing your own tools.
In a nutshell
- Create a project to hold your analyses on the Seven Bridges Platform.
- Add files to your project and supply their metadata to prepare them for analyses. Don't forget to add reference files!
- Add and edit a public workflow (Whole Exome Sequencing - BWA + GATK 4.0) to run your whole exome sequencing analysis.
- Set up your task on the DRAFT task page by selecting inputs and reference files.
- The Outputs page displays the results of your task.
Suggested pages:
Troubleshoot a failed task
API Quickstart
Seven Bridges Platform tutorials
Updated 3 months ago
